‘TBRDist’ is an R package to compare unrooted phylogenetic trees using the SPR, TBR and Replug distances.
Loading trees
This document assumes familiarity with the process of loading trees into R.
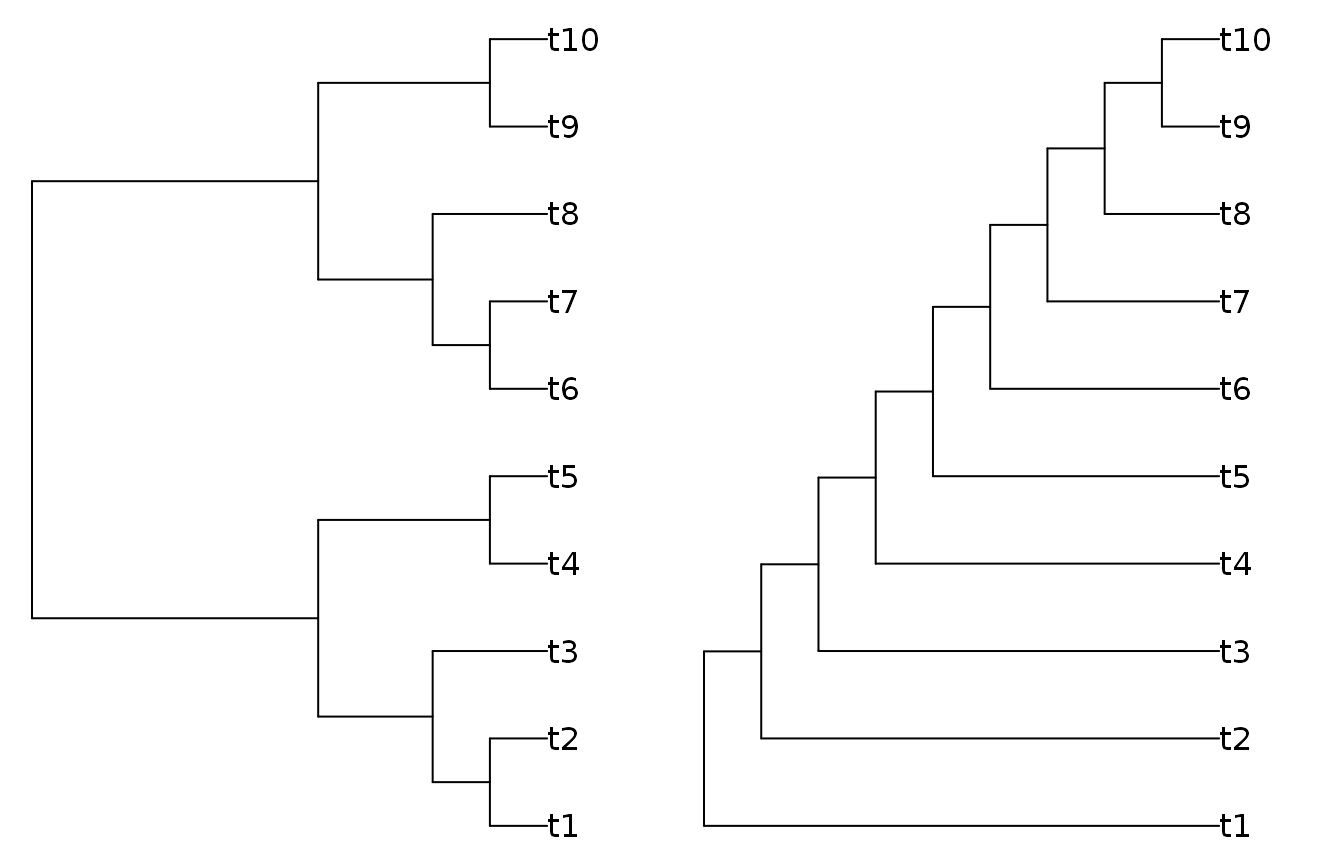
We’ll work with some simple trees generated using the ‘TreeTools’ package.
library('TreeTools', quietly = TRUE, warn.conflicts = FALSE) tree1 <- BalancedTree(10) tree2 <- PectinateTree(10) origPar <- par(mfrow = 1:2, mar = rep(0.2, 4)) # Set up plotting area plot(tree1) plot(tree2)
Calculating distances
After installing TBRDist (install.packages('TBRDist')), load the package into R with
To generate an approximate SPR distance between two unrooted trees, use:
USPRDist(tree1, tree2)
## [1] 2For Replug distances, it’s:
ReplugDist(tree1, tree2)
## [1] 2For TBR distances, use:
TBRDist(tree1, tree2, exact = TRUE)
## [1] 1When calculating an exact TBR distance, we receive information on the maximum agreement forest for free (i.e. with no extra processing cost):
TBRDist(tree1, tree2, exact = TRUE, maf = TRUE)
## $tbr_exact
## [1] 1
##
## $maf_1
## [1] "(t4,t5,((t1,t2),t3)); ((t9,t10),t8,(t7,t6));"
##
## $maf_2
## [1] "(((t1,t2),t3),t4,t5); ((t6,t7),(t9,t10),t8);"Once trees have more than about a dozen tips, it becomes slow to calculate the exact distance. In the interests of speed, we may wish to approximate the value of the TBR distance:
TBRDist(tree1, tree2, exact = FALSE)
## $tbr_min
## [1] 1
##
## $tbr_max
## [1] 3Comparing multiple trees
To avoid multiple calls, each function can also be used to compare lists of trees (or multiPhylo objects) against a single tree:
## $tbr_exact
## [1] 0 1
##
## $maf_1
## [1] "((((t1,t2),t3),(t4,t5)),(t9,t10),((t6,t7),t8));"
## [2] "(t4,t5,((t1,t2),t3)); ((t9,t10),t8,(t7,t6));"
##
## $maf_2
## [1] "((((t1,t2),t3),(t4,t5)),(t9,t10),((t6,t7),t8));"
## [2] "(((t1,t2),t3),t4,t5); ((t6,t7),(t9,t10),t8);"Or against each corresponding entry in a second list:
## [1] 2 2 0Citation
If you use ‘TBRDist’ in your research, please cite:
- Chris Whidden and Frederick A. Matsen IV (2017). Calculating the Unrooted Subtree-Prune-and-Regraft Distance. eprint arXiv:1511.07529.
Optionally, a citation to the ‘TBRDist’ R package would be welcome:
- Smith (2019). TBRDist: Calculate SPR and TBR distance between unrooted phylogenetic trees. doi:10.5281/zenodo.3548333