Treeness (also termed stemminess) is the proportion of a tree's length found on internal branches (Lanyon 1988) . Insofar as external branches do not contain phylogenetic (grouping) signal, trees with a high treeness can be interpreted as containing a higher signal:noise ratio (Phillips and Penny 2003-08) .
Arguments
- tree
A tree of class
phylo, or a list of trees of classlistormultiPhylo.
References
Lanyon SM (1988).
“The Stochastic Mode of Molecular Evolution: What Consequences for Systematic Investigations?”
The Auk, 105(3), 565–573.
doi:10.1093/auk/105.3.565
.
Phillips MJ, Penny D (2003-08).
“The Root of the Mammalian Tree Inferred from Whole Mitochondrial Genomes.”
Molecular Phylogenetics and Evolution, 28(2), 171–185.
doi:10.1016/S1055-7903(03)00057-5
.
See also
Other tree properties:
Cherries(),
ConsensusWithout(),
LongBranch(),
MatchEdges(),
NSplits(),
NTip(),
NodeNumbers(),
PathLengths(),
SplitsInBinaryTree(),
TipLabels(),
TreeIsRooted()
Examples
lowTree <- BalancedTree(6, lengths = c(1, 1, 4, 4, 4, 1, 1, 4, 4, 4))
plot(lowTree)
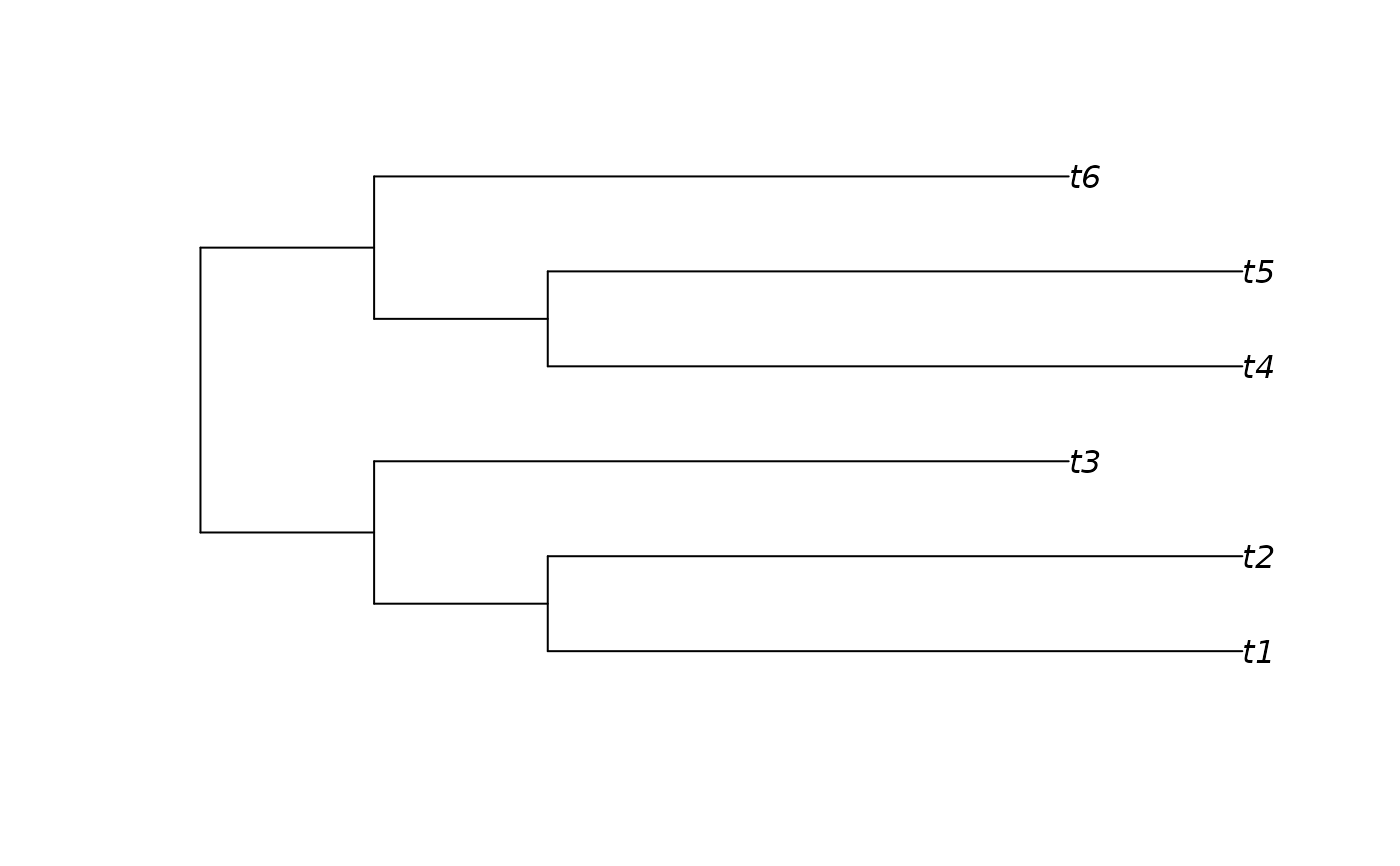 Treeness(lowTree)
#> [1] 0.1428571
highTree <- BalancedTree(6, lengths = c(6, 6, 1, 1, 1, 6, 6, 1, 1, 1))
plot(highTree)
Treeness(lowTree)
#> [1] 0.1428571
highTree <- BalancedTree(6, lengths = c(6, 6, 1, 1, 1, 6, 6, 1, 1, 1))
plot(highTree)
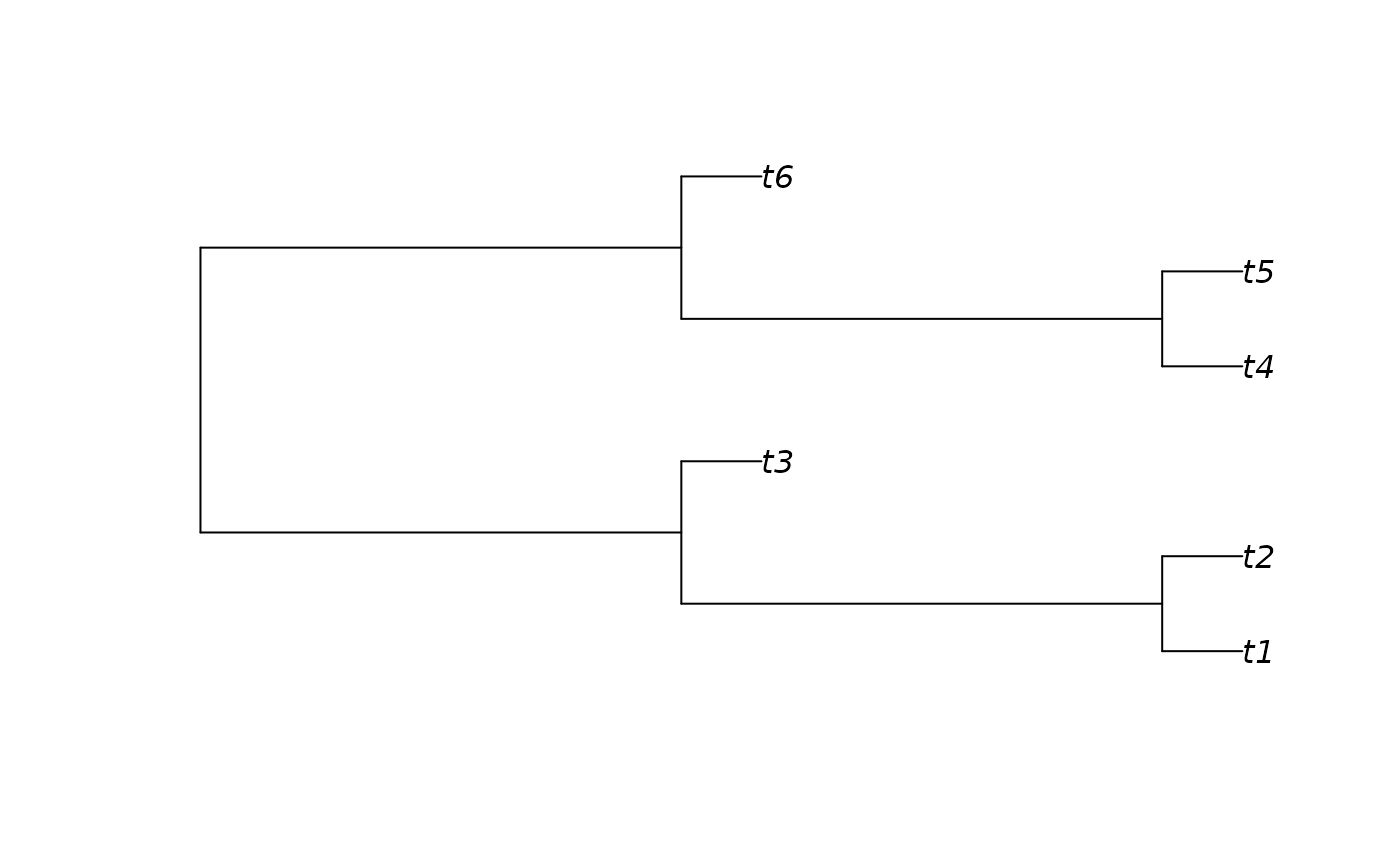 Treeness(c(lowTree, highTree))
#> [1] 0.1428571 0.8000000
Treeness(c(lowTree, highTree))
#> [1] 0.1428571 0.8000000