2 Fitch parsimony
Parsimony search with the Fitch (1971) algorithm was conducted in TNT v1.5 (Goloboff & Catalano, 2016) using Ratchet and tree drifting heuristics (Goloboff, 1999; Nixon, 1999), repeating the search until the optimal score had been hit by 1500 independent searches:
xmult:rat25 drift25 hits 1500 level 4 chklevel 5;
Searches were conducted under equal weights and results saved to file:
piwe-; xmult; /* Conduct search with equal weighting */
tsav *TNT/ew.tre;sav;tsav/; /* Save results to file */
Node support was estimated by calculating the proportion of jackknife replicates in which each group occurred, using 5000 symmetric resampling iterations, following the recommendations of Kopuchian & Ramírez (2010) and Simmons & Freudenstein (2011).
var: nt; /* Define a variable to track tree address */
nelsen *; /* Generate strict consensus tree */
set nt ntrees; ttag=; /* Prepare for resampling */
resample=sym 5000 frequency from ‘nt’; /* Symmetric resampling, counting frequencies */
log TNT/ew.sym; ttag/; log/; /* Write results to log */
keep 0; ttag-; hold 10000; /* Clear memory */
Further searches were conducted under extended implied weighting (Goloboff, 1997, 2014), under the concavity constants 3, 4.5, 7, 10.5, 16 and 24:
xpiwe=; /* Enable extended implied weighting */
piwe=3; xmult; /* Conduct analysis at k = 3 */
tsav *TNT/xpiwe3.tre; sav; tsav/; /* Save results to file */
nelsen *; set nt ntrees; ttag=; /* Prepare for resampling */
resample=frequency from ‘nt’; /* Symmetric resampling */
log TNT/xpiwe3.sym; ttag/; log/; /* Write results to log */
keep 0; ttag-; hold 10000; /* Clear memory */
/* Repeat this block for each value of k */
Results can be replicated by:
- Downloading the data in TNT format.
- Saving the script above to the same directory, with the filename
tnt.run. - Opening TNT, typing
piwe=before you load the downloaded dataset (to enable extended implied weighting), then typingtntinto the command box to run the script.
We acknowledge the Willi Hennig Society for their sponsorship of the TNT software.
2.1 Results
Optimal trees can be downloaded in TNT Newick format from github.com/ms609/hyoliths/tree/master/TNT.
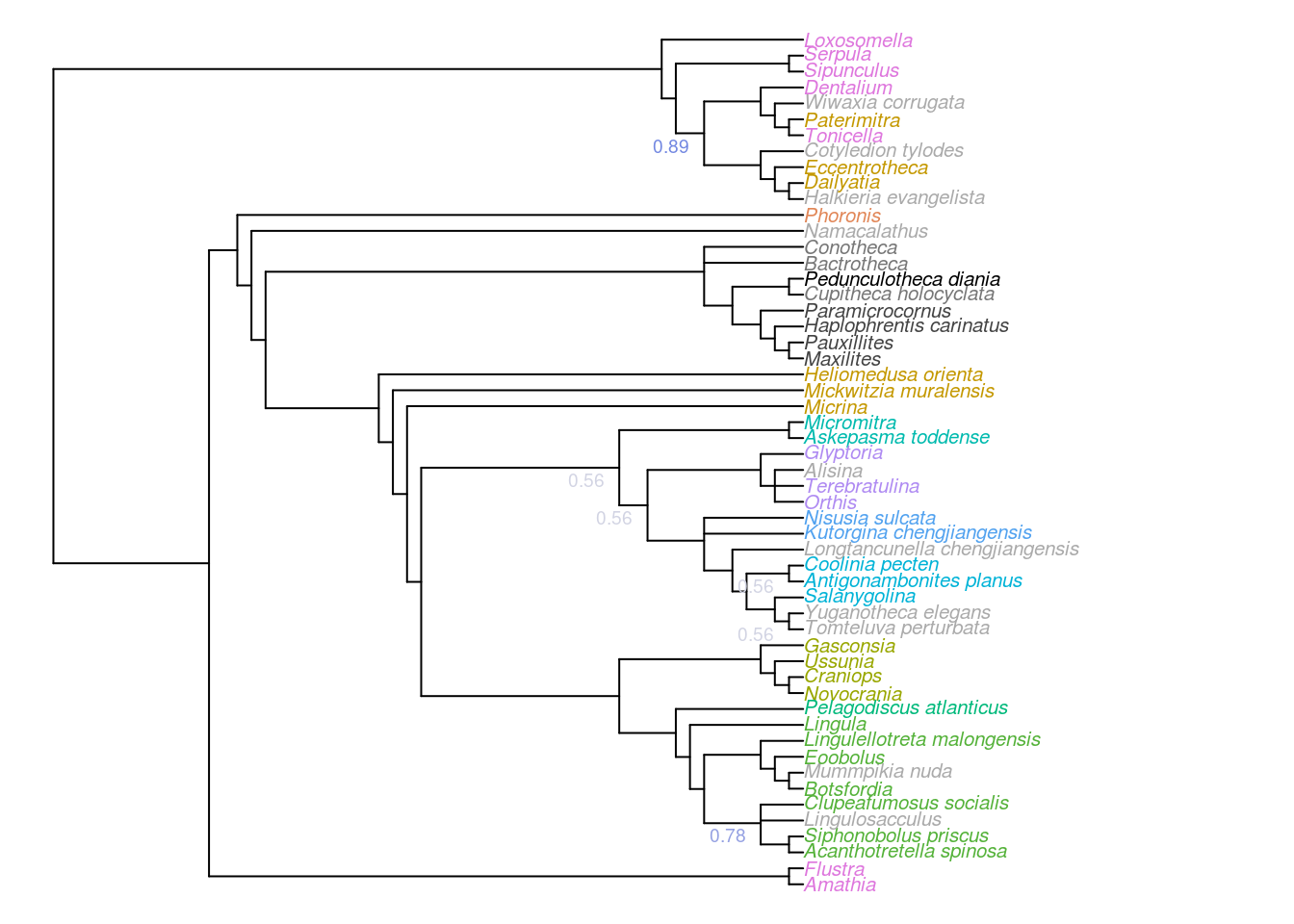
Figure 2.1: Majority-rule consensus of all trees that are optimal under implied weights. Node labels denote, where less than 100%, the frequency of each node in the set of all optimal trees.
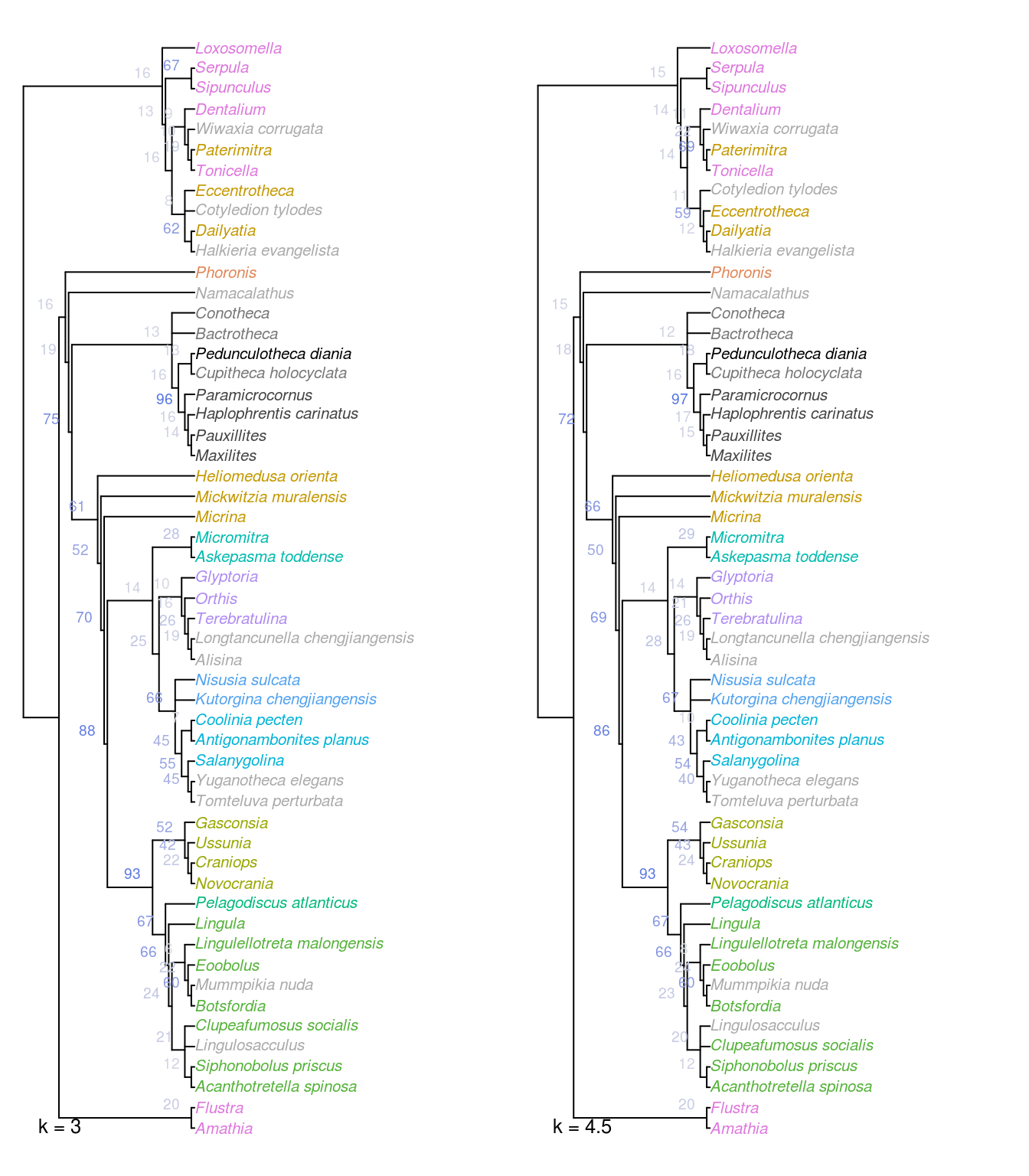
Figure 2.2: Strict consensus of all optimal trees under Fitch parsimony with implied weighting at k = 3 and 4.5. Nodes labelled with jackknife frequencies (%).
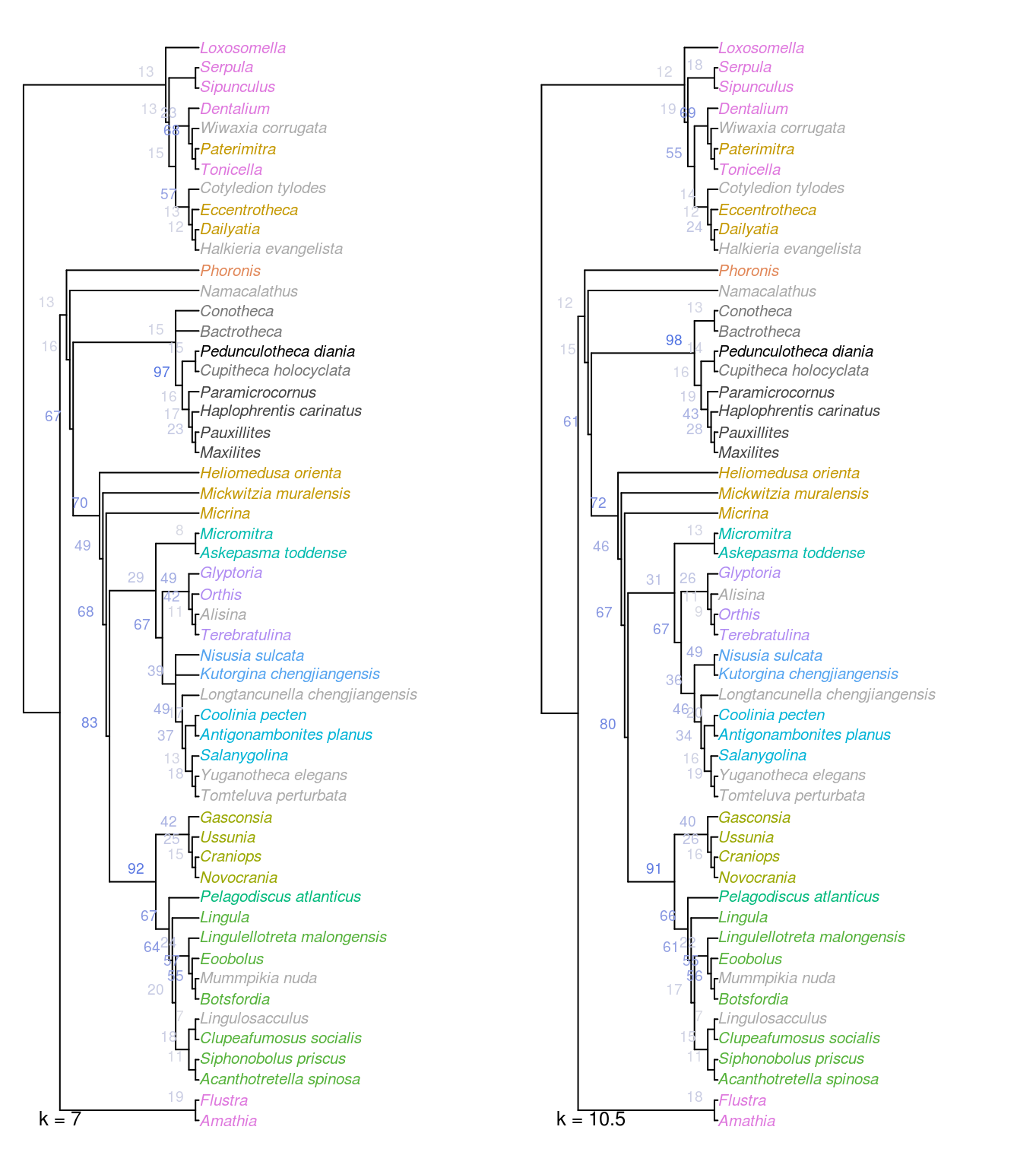
Figure 2.3: Strict consensus of all optimal trees under Fitch parsimony with implied weighting, at k = 7 and 10.5.
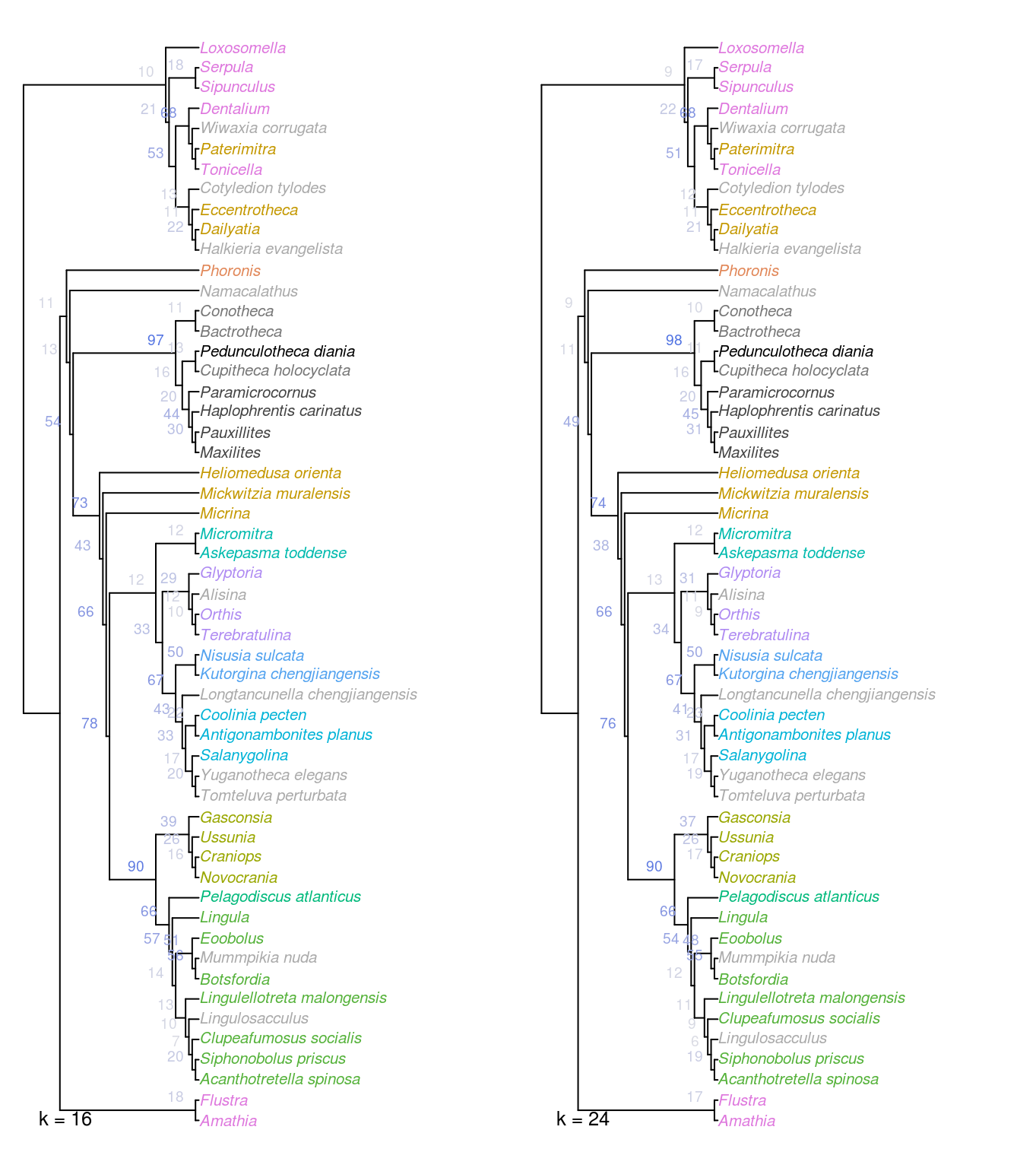
Figure 2.4: Strict consensus of all optimal trees under Fitch parsimony with implied weighting, at k = 16 and 24.
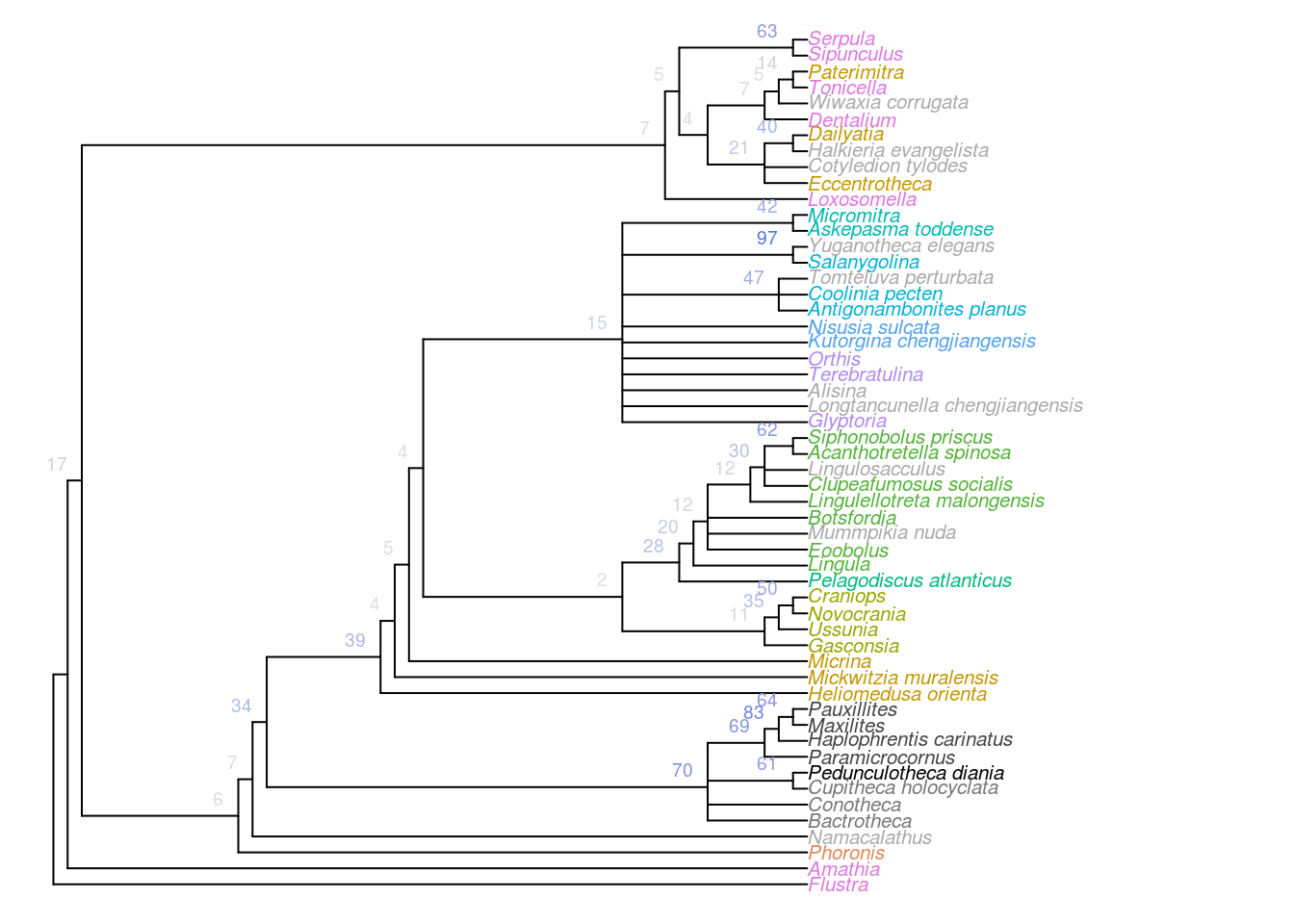
Figure 2.5: Consensus of all trees that are optimal under equally weighted Fitch parsimony. Nodes labelled with jackknife frequencies (%).
References
Fitch, W. M. (1971). Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Biology, 20(4), 406–416. doi:10.1093/sysbio/20.4.406
Goloboff, P. A., & Catalano, S. A. (2016). TNT version 1.5, including a full implementation of phylogenetic morphometrics. Cladistics, 32(3), 221–238. doi:10.1111/cla.12160
Goloboff, P. A. (1999). Analyzing large data sets in reasonable times: solutions for composite optima. Cladistics, 15(4), 415–428. doi:10.1006/clad.1999.0122
Nixon, K. C. (1999). The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics, 15(4), 407–414. doi:10.1111/j.1096-0031.1999.tb00277.x
Kopuchian, C., & Ramírez, M. J. (2010). Behaviour of resampling methods under different weighting schemes, measures and variable resampling strengths. Cladistics, 26(1), 86–97. doi:10.1111/j.1096-0031.2009.00269.x
Simmons, M. P., & Freudenstein, J. V. (2011). Spurious 99% bootstrap and jackknife support for unsupported clades. Molecular Phylogenetics and Evolution, 61(1), 177–191. doi:10.1016/j.ympev.2011.06.003
Goloboff, P. A. (1997). Self-weighted optimization: tree searches and character state reconstructions under implied transformation costs. Cladistics, 13(3), 225–245. doi:10.1111/j.1096-0031.1997.tb00317.x
Goloboff, P. A. (2014). Extended implied weighting. Cladistics, 30(3), 260–272. doi:10.1111/cla.12047