Collapses specified nodes or edges on a phylogenetic tree, resulting in polytomies.
Usage
CollapseNode(tree, nodes)
# S3 method for class 'phylo'
CollapseNode(tree, nodes)
CollapseEdge(tree, edges)Arguments
- tree
A tree of class
phylo.- nodes, edges
Integer vector specifying the nodes or edges in the tree to be dropped. (Use
nodelabels()oredgelabels()to view numbers on a plotted tree.)
Value
CollapseNode() and CollapseEdge() return a tree of class phylo,
corresponding to tree with the specified nodes or edges collapsed.
The length of each dropped edge will (naively) be added to each descendant
edge.
See also
Other tree manipulation:
AddTip(),
ConsensusWithout(),
DropTip(),
ImposeConstraint(),
KeptPaths(),
KeptVerts(),
LeafLabelInterchange(),
MakeTreeBinary(),
Renumber(),
RenumberTips(),
RenumberTree(),
RootTree(),
SortTree(),
Subtree(),
TipTimedTree(),
TrivialTree
Examples
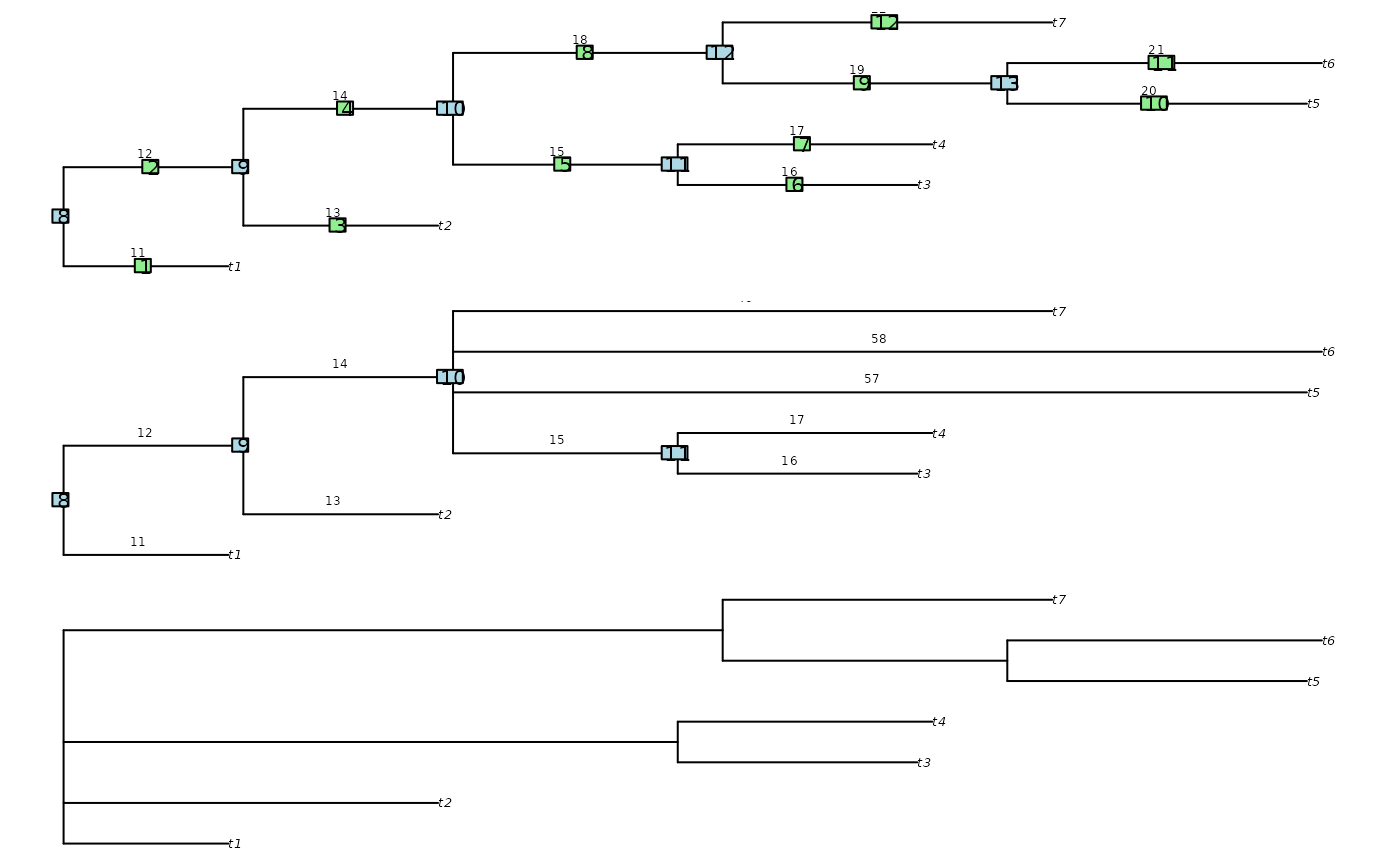
oldPar <- par(mfrow = c(3, 1), mar = rep(0.5, 4))
tree <- as.phylo(898, 7)
tree$edge.length <- 11:22
plot(tree)
nodelabels()
edgelabels()
edgelabels(round(tree$edge.length, 2),
cex = 0.6, frame = "n", adj = c(1, -1))
# Collapse by node number
newTree <- CollapseNode(tree, c(12, 13))
plot(newTree)
nodelabels()
edgelabels(round(newTree$edge.length, 2),
cex = 0.6, frame = "n", adj = c(1, -1))
# Collapse by edge number
newTree <- CollapseEdge(tree, c(2, 4))
plot(newTree)
 par(oldPar)
par(oldPar)